A research team from CPS-ZJU published an article in Nature Communications: Tokenized Drug Design Based on Large Language Models
2025-05-16 | 药学院英文网
In recent years, large language models (LLMs) have made significant progress in the application of drug design. However, the existing methods often struggle to effectively integrate the three-dimensional structure of molecules. Therefore, developing a chemical large model that is applicable to all drug design scenarios and is easy to integrate with existing general large language models has become a key issue that needs to be urgently addressed at present.
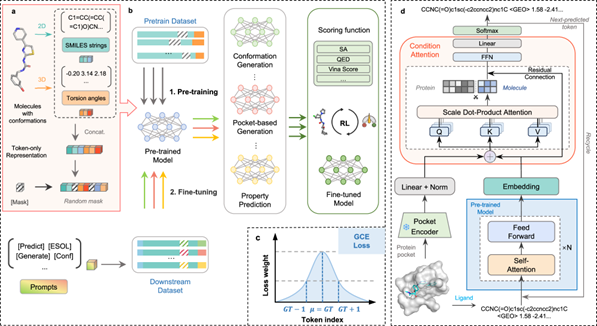
On May 13, 2025, the team of Tingjun Hou, Changyu Xie and Yu Kang from CPS-ZJU published a paper titled Token-Mol 1.0: Tokenized Drug Design with Large Language Models in Nature Communications. This research proposed a three-dimensional drug design model called Token-Mol, which only uses word tokens to encode two-dimensional and three-dimensional structure information as well as molecular properties. Token-Mol is based on the decoder architecture of the transformer and adopts causal masking training. It introduces a Gaussian cross-entropy loss function (GCE) tailored for regression tasks, thereby achieving excellent performance in multiple downstream applications. This model outperforms existing methods in multiple downstream tasks: in the molecular conformation task, the performance on two datasets has improved by more than 10% and 20% respectively, and in the property prediction task, it outperforms other models based solely on word tokens by 30%. In the pocket-based molecular generation, compared with other expert models, Token-Mol increases drug similarity and synthesis accessibility by approximately 11% and 14% respectively, and in terms of generation speed, it is 35 times faster than the expert model based on diffusion models. In the generation of scenarios simulating real-world situations, when combined with reinforcement learning, the model can optimize the affinity and drug similarity of the designed molecules in one step.
NEWS
-
10
2025.12
-
27
2025.11
-
25
2025.11
-
03
2025.11
-
30
2025.10
-
29
2025.10
