CPS-ZJU research team reports spatial transcriptome data simulation framework scCube in Nature Communications
2024-06-15 | 药学院英文网
The emergence and rapid development of spatially resolved transcriptomics (SRT) sequencing technologies has provided unprecedented opportunities to understand the cellular composition, molecular structure and functional details of tissues at the spatial level. With the availability of SRT data, computational tools for a variety of downstream analysis tasks have become increasingly sophisticated. While these computational methods are often based on reasonable assumptions, it is difficult to benchmark and evaluate their performance without a gold standard. Currently, a widely used approach is to utilise simulated data. However, existing simulated SRT data are biased, which seriously affects the accuracy of method evaluation and validation. In addition, the lack of reproducibility in the description of most current simulation steps may hinder the potential use of these simulation methods by other researchers. Therefore, there is an urgent need for a generic simulation framework for SRT data that is capable of simulating unbiased, independent, reproducible, and with a variety of platforms to better facilitate the development of various downstream analytical tools.

On 12 June 2024, the team of Prof. Yiyu Cheng and Prof. Xiaohui Fan from the College of Pharmaceutical Sciences and National Key Laboratory of Modern Chinese Medicine jointly published a research paper entitled “Simulating multiple variability in spatially resolved transcriptomics with scCube” in the international journal Nature Communications. They developed a simulation framework called scCube to simulate multiple spatial variability in SRT data and generate unbiased simulated SRT data, which enables researchers to systematically investigate different types of SRT computational methods more easily and accurately. types of SRT computational methods for systematic and comprehensive benchmarking and evaluation.
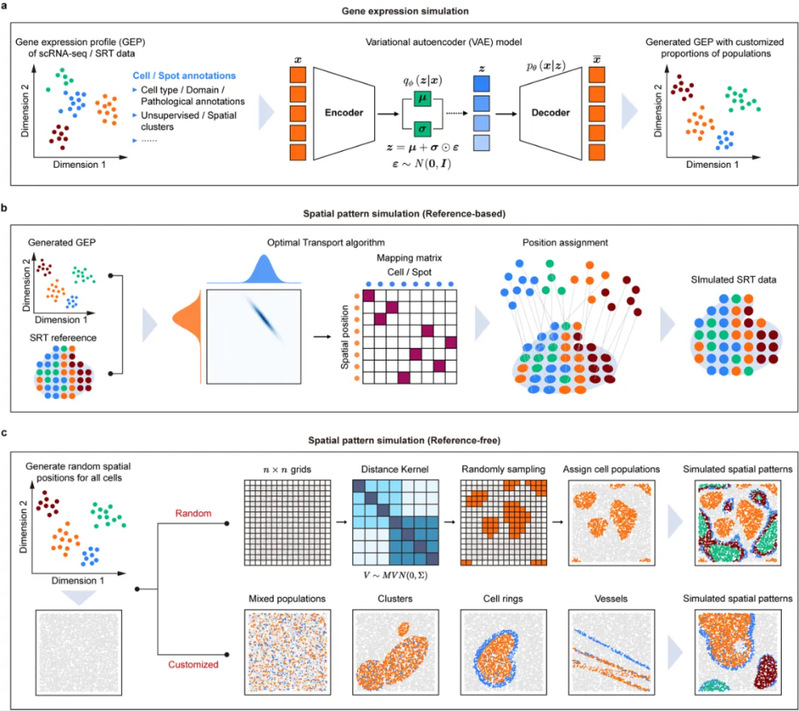
Figure 1. Schematic diagram of the principle of the scSpace algorithm
The first authors of this paper are Jingyang Qian, a PhD student, and Hudong Bao, an undergraduate student of CPS-ZJU. Prof. Yiyu Cheng and Prof. Xiaohui Fan from College of Pharmaceutical Sciences and National Key Laboratory of Modern Chinese Medicine , were the co-corresponding authors of this article.
Original link:https://www.nature.com/articles/s41467-024-49445-0
NEWS
-
10
2025.12
-
27
2025.11
-
25
2025.11
-
03
2025.11
-
30
2025.10
-
29
2025.10
