CPS-ZJU research team reports a new method for inferring drug effector cell types based on single-cell transcriptome data in Cell Reports Medicine
2024-05-30 | 药学院英文网
It is well known that there are significant differences in the response of different cell types to drugs in tissues. With the help of single-cell transcriptome sequencing technology, it is possible to analyse the expression of different RNA molecules within a single cell, thus resolving and typing heterogeneous cell populations. However, it remains a challenge to identify key cell types for drug effects in single-cell data. For example, many drugs are efficacious, but it is unclear by which cell populations these drugs function by affecting. Therefore, it is important to elucidate the mechanism of action of drugs (especially complex drugs such as traditional Chinese medicines) by revealing which cells are drug effector cells to promote precision therapy.

On 15 May 2024, the team of Prof. Yi Wang and Prof. Xiaohui Fan from CPS-ZJU published the paper “scRank infers drug-responsive cell types from untreated scRNA-seq data using a target-perturbed gene regulatory network” online in Cell Reports Medicine. The paper uses a target-perturbed gene regulatory network (tpGRN) to model drug perturbation by integrating biological networks and drug target information, and achieves the inference of drug-responsive cell types from untreated single-cell transcriptome data only.
The research team used scRank to successfully analyse the mechanism of action of tanshinones in the blood-activating and blockage-eliminating traditional Chinese medicine Danshen, which has complex targets, and deduced the macrophage subpopulations that are mainly inhibited by tanshinone IIA during the process of myocardial infarction alleviation. The study further demonstrated that scRank can reveal the potential molecular targets of traditional Chinese medicine and natural product molecules with complex targets. By inputting one of the known targets of tanshinone IIA and the target macrophage subpopulation of tanshinone IIA, the research team was able to sequence the gene nodes of the macrophage subpopulation biological network, and identified other potential targets of tanshinone IIA. In summary, scRank can accurately infer the effector cell types and potential targets of drugs from unprocessed scRNA-seq data, which provides an important tool for understanding the mechanism of drug action at the single-cell level, and can provide a new research strategy for the identification of efficacy of complex systems of traditional Chinese medicines, and for precision therapy, etc. The scRank tool has been open sourced to GitHub.
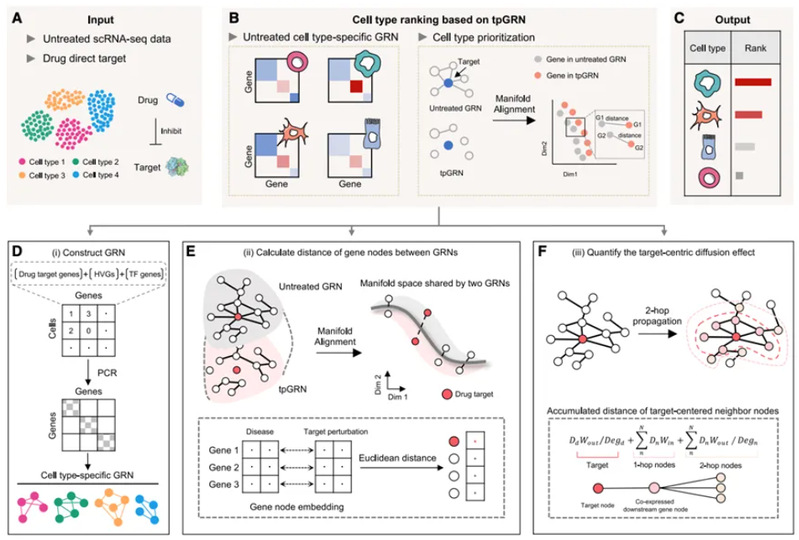
Figure 1. Schematic diagram of the principle of the scRank algorithm
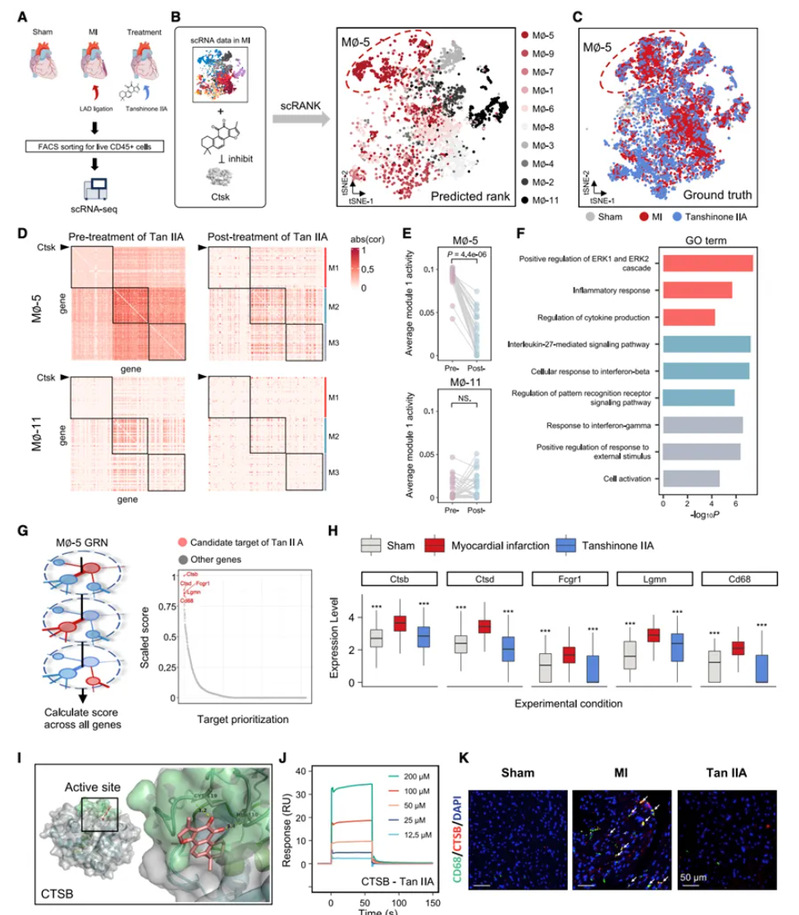
Figure 2. Identification of effector macrophage subpopulations and potential targets of tanshinone IIA using scRank
The first authors of this article are Chengyu Li, a PhD from CPS-ZJU, and Xin Shao, a researcher of ,Innovation Center of Yangtze River Delta, Zhejiang University. Professor Yi Wang, Professor Xiaohui Fan, and Researcher Shao Xin from School of Pharmacy and National Key Laboratory of Modern Chinese Medicine are the corresponding authors of this paper.
Original link:https://www.sciencedirect.com/science/article/pii/S266637912400260X?via%3Dihub
NEWS
-
10
2025.12
-
27
2025.11
-
25
2025.11
-
03
2025.11
-
30
2025.10
-
29
2025.10
