CPS-ZJU scientist report a novel representation learning-based deep clustering framework for accurate visual inspection in virtual screening
2023-05-07 | 药学院英文网
In April 2023, Professor Xiaowu Dong and research fellow Jinxin Che's team from the College of Pharmaceutical Sciences at Zhejiang University, along with Professor Wu Jian from the School of Medicine, published a paper titled ClusterX: a novel representation learning-based deep clustering framework for accurate visual inspection in virtual screening in Briefings in Bioinformatics. They proposed a new method for protein-ligand complex deep clustering to assist visual inspection in virtual screening. This method introduces a protein-ligand complex representation learning network to improve molecular representations for clustering. The learned molecular representations are input into the clustering module for joint optimization with the graph representation learning module using a common optimization strategy during the training process.

Molecular clustering analysis has been developed to facilitate visual inspection in the process of structure-based virtual screening. However, traditional methods based on molecular fingerprints or molecular descriptors limit the accuracy of selecting active hit compounds, which may be attributed to the lack of representations of receptor structural and protein–ligand interaction during the clustering. Here, a novel deep clustering framework named ClusterX is proposed to learn molecular representations of protein–ligand complexes and cluster the ligands. In ClusterX, the graph was used to represent the protein–ligand complex, and the joint optimisation can be used efficiently for learning the cluster-friendly features. Experiments on the KLIFs database show that the model can distinguish well between the binding modes of different kinase inhibitors. To validate the effectiveness of the model, the clustering results on the virtual screening dataset further demonstrated that ClusterX achieved better or more competitive performance against traditional methods, such as SIFt and extended connectivity fingerprints. This framework may provide a unique tool for clustering analysis and prove to assist computational medicinal chemists in visual decision-making.
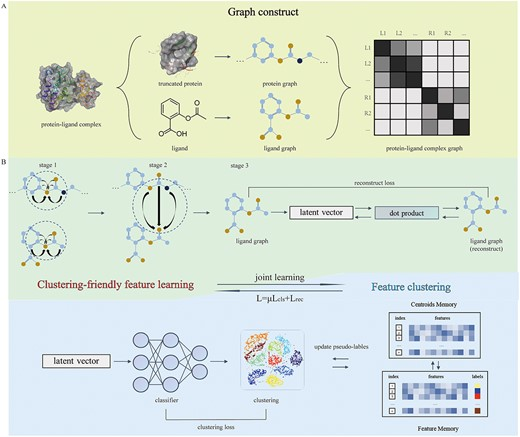

CPS-ZJU is the first affiliation of this paper, and Sikang Chen, a master student, is the first author. Professor Xiaowu Dong, Professor Jian Wu, and Associate Researcher Jinxin Che are the co-corresponding authors. This research is supported by the Zhejiang University Institute of Intelligent Innovation Medicines platform and by projects such as the National Natural Science Foundation, the Outstanding Youth Project of Zhejiang Province Natural Science Foundation, and the Zhejiang Province Key R&D Plan.
Link: https://academic.oup.com/bib/advance-article/doi/10.1093/bib/bbad126/7107930
Translator: Mingyang Zhou
Editor: Yichen Zhu
NEWS
-
10
2025.12
-
27
2025.11
-
25
2025.11
-
03
2025.11
-
30
2025.10
-
29
2025.10
